The West African Centre for Cell Biology of Infectious Pathogens (WACCBIP) has successfully concluded an end-to-end two-week genomics workshop (both ‘wet’ lab and downstream ‘dry lab’ workflows) on "Combatting AMR: Genome Sequencing and Bioinformatics."
Taking place from February 19 to March 1, 2024, this event brought together a diverse community of experts, researchers, including students and Postdocs and enthusiasts determined to make significant strides in the battle against antimicrobial resistance (AMR).

The workshop facilitators were Dr Amenga-Etego Lucas, Dr Morang'a Collins Misita, Dr Bah Saikou Y, Dr Abiola Isawunmi, Dr Duodu Samuel, Ms Amu Thompson Kukua, Ms Mwongeli Ngoi Joyce, Mr Appiah Vincent, and Mr Dzabeng Francis.
Opening the workshop, Dr Amenga-Etego gave an in-depth talk on Genomics for AMR studies, where he explored the role of genomics in AMR studies and laid the foundation for genomic approaches to understanding and combating AMR.
Highlights of other topics delivered to both cohorts included:
Mechanisms of AMR: A comprehensive exploration of the mechanisms behind antimicrobial resistance, providing insights into how pathogens evolve and develop resistance to antimicrobial agents.
AMR for One Health: Emphasizing the interconnectedness of human, animal, and environmental health, this session underscored the importance of a holistic approach to combating AMR.
Public Databases for AMR: An overview of key public databases for AMR, coupled with practical sessions on AMR detection and retrieval of information about AMR genes for both cohorts.

Evolution of Drug Resistance: A fascinating look into the evolution of drug resistance in an ancient TB pathogen, exploring the emergence, fixation, spread, and reconstruction of whole genome sequences.
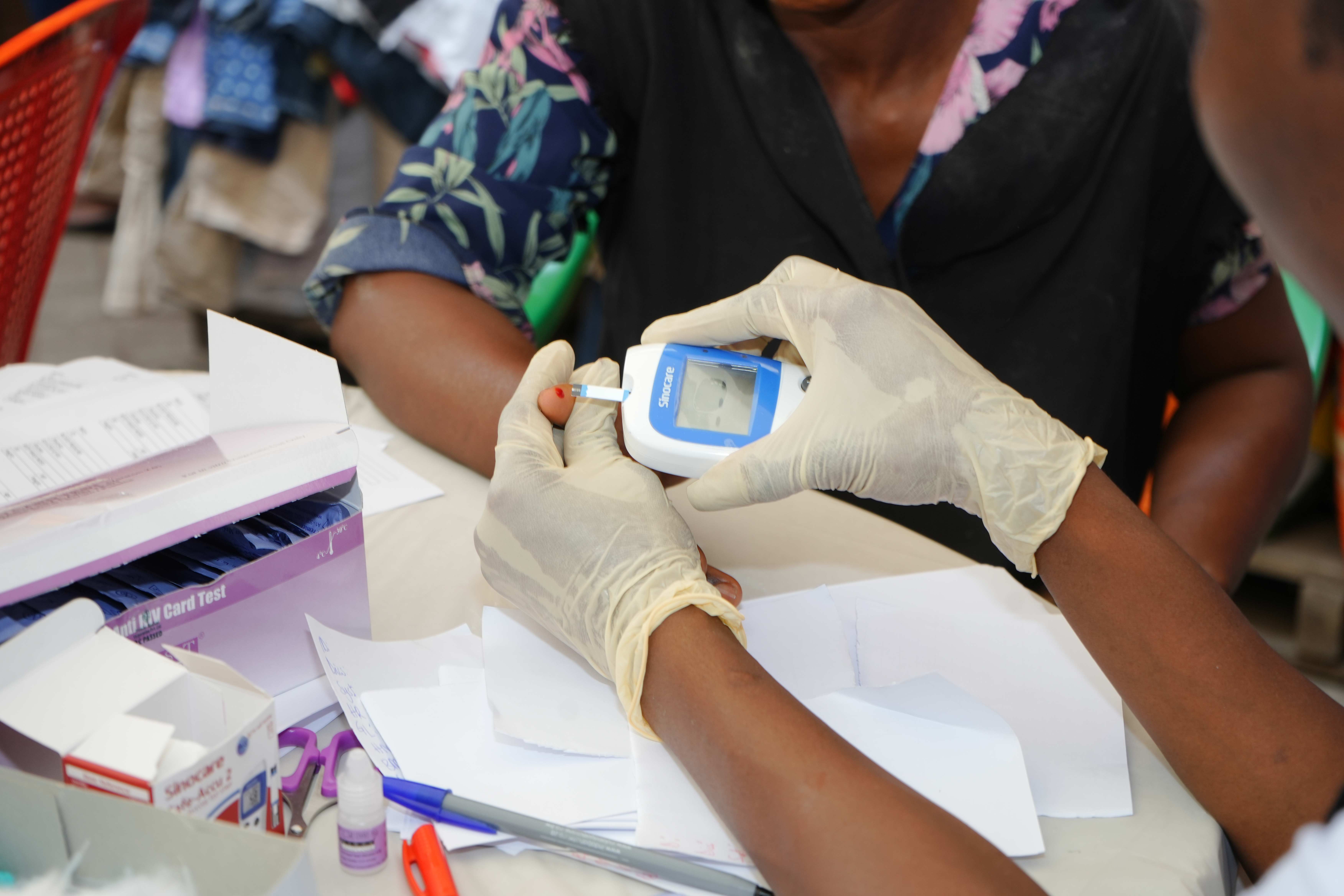
Introduction to Next-Generation Sequencing (NGS) workflows: A practical session introducing participants in the wet lab cohort to the fundamentals of NGS, from library prep to sequencing, including laboratory safety tips and data management strategies and pitfalls.

The ‘dry lab’ participants from diverse skill levels were treated to:
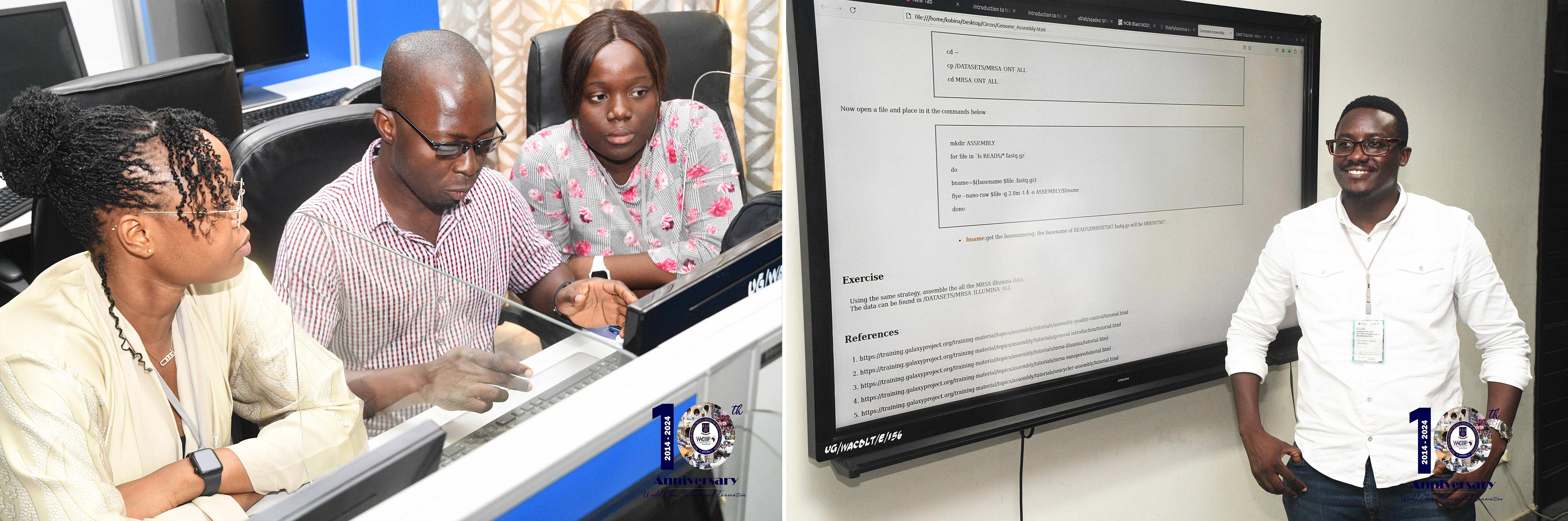
Linux Basics: A hands-on exploration of Linux, covering basic commands, file and directory management, and essential skills for researchers working in bioinformatics.
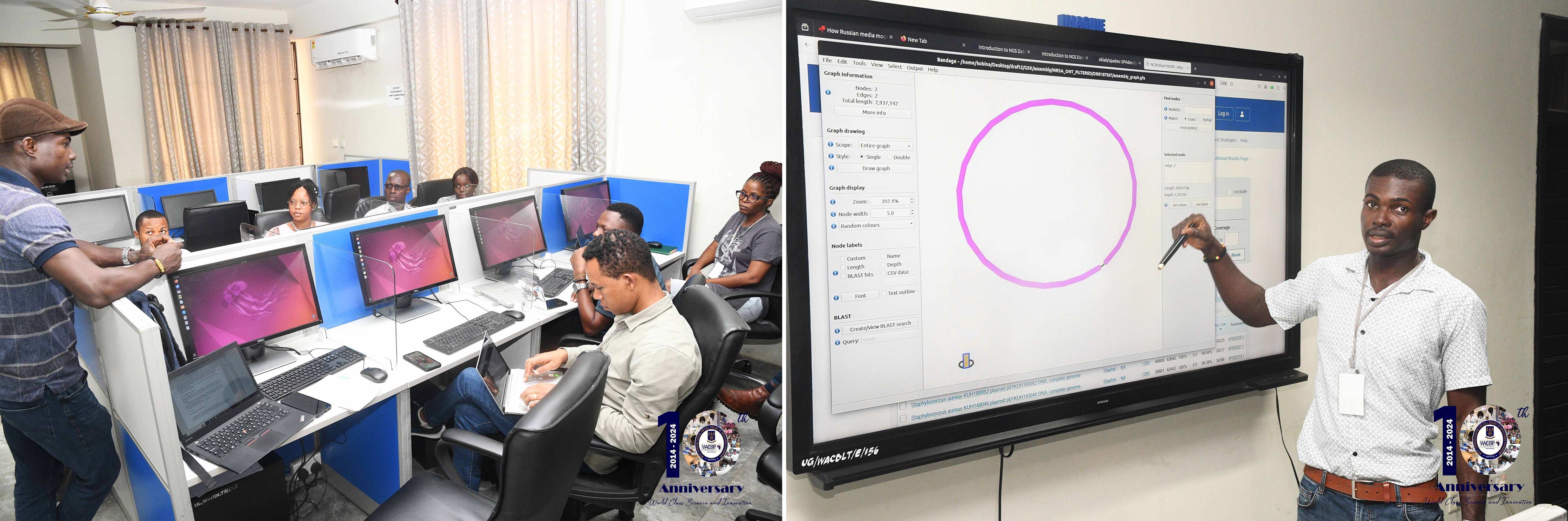
NGS Data Analysis: Participants delved into the world of NGS data types, read trimming and filtering, genome assembly of short and long reads, and hybrid genome assembly.
Bacterial Genome Annotation: Hands-on training in genome annotation using tools like Prokka, PlasmidFinder, STARAMR, and ABRicate, including techniques to identify plasmids and AMR genes.
Phylogenetic Analysis: The workshop concluded with practical sessions on estimating phylogenetic trees using RaxML, multiple sequence alignment, and visualizing phylogenetic trees using iTOL.
The participants, having completed the intensive workshop, received certificates and WACCBIP-branded souvenirs during the closing ceremony conducted by WACCBIP Director, Professor Gordon A. Awandare. The workshop not only imparted knowledge but also fostered collaboration and a renewed commitment to combating AMR on multiple fronts. Other highlights included a visit to Ghana’s founding president Dr Kwame Nkrumah’s mausoleum, and Ghana’s Independence Square among others.

The workshop, which was supported by GSK has set a ripple in motion to build and expand the capacity for AMR genomics in Africa to ensure a new era in the fight against antimicrobial resistance, empowering researchers and professionals to employ advanced genomics and bioinformatics strategies to tackle one of the most pressing global health challenges of our time.