The West African Centre for Cell Biology of Infectious Pathogens (WACCBIP), in collaboration with the Institut de Recherche pour le Développement (IRD) France, and with support from Wellcome Connecting Science has organised a virtual Helminths Bioinformatics course from Monday, November 08 to Friday, November 12, 2021. The week-long course, which was held via Zoom brought together over 100 participants from across Africa to equip them with the requisite skills to access, analyse, and display large-scale helminth genomic data.
The course organisers and instructors were Mr Vincent Appiah, High-Performance Computing Manager at WACCBIP, Mr Collins M. Morang’a, a Bioinformatician at WACCBIP, and Dr Lucas Amenga-Etego – a Senior Research Fellow at WACCBIP. They were supported by Dr Nicaise Tuikue Ndam, Head of the Benin-Ghana-Nigeria-Togo cluster of IRD; Dominic Amuzu, a PhD Research Fellow at WACCBIP; Dr Samuel Kwofie, Senior Lecturer at the Department of Biomedical Engineering, University of Ghana; Dr Thomas Otto –Senior Lecturer at the University of Glasgow, Dr Alice Matimba – Acting Head of Courses and Global Training at Wellcome Connecting Science; and Dr Steve Doyle – a Molecular and Computational Biologist at the Wellcome Sanger Institute.
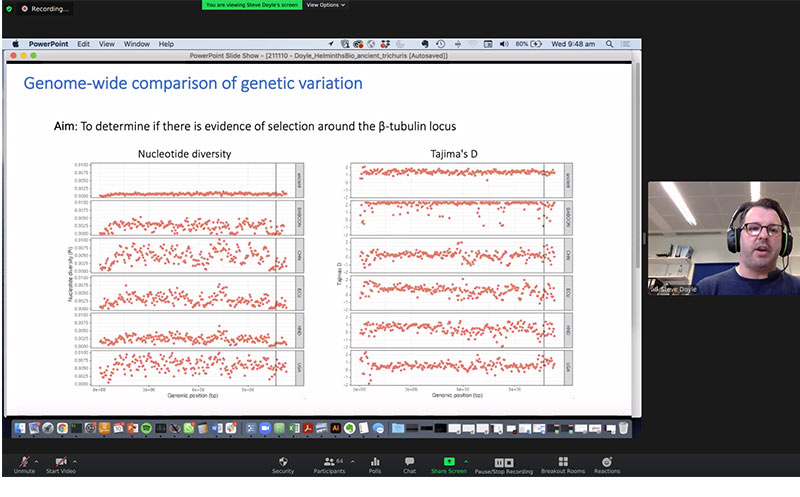
Dr Steve Doyle delivers a presentation on Genome-wide Comparison of Genetic Variation.
Helminths are parasitic worms that feed on a living host to gain nourishment and protection while causing poor nutrient absorption, weakness, and disease. Some of these worms live in the intestines and are referred to as intestinal parasites. Helminth Bioinformatics, therefore, entails the study of genetic characteristics of these worm-like parasites using computational tools and techniques including DNA sequence alignments and variations (Genomics), navigation of biological databases, finding large biological datasets, analysis of gene information (Transcriptomics).
Dr Thomas Otto introduces participants to Transcriptomics
Some of the topics covered during the training included the General Concept of Bioinformatics and its applications presented by Dr Lucas Amenga-Etego; Population Genomics by Dr Steve Doyle where participants were put into groups and made to analyse genetic variation using R, a software used for cleaning, analysing and graphing data. Participants were also introduced to the use of the Linux command line, a text interface, as well as the Artemis software, a genome browser used to visualise the variations. The practical sessions offered participants the opportunity to try their hands on RNA-seq transcriptomics on R and Linux. The last session was on building partnerships and collaborations, where participants on building partnerships and how to collaborate with others in a mutually beneficial manner.
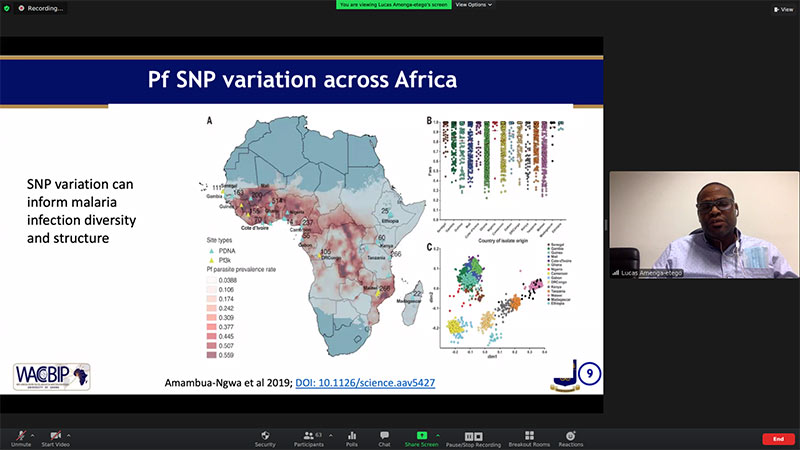
Dr Lucas Amenga-Etego delivers a lecture during the course.
Some participants took turns to express their views after the week-long course. Dr Stéphane Koffi, a Researcher with Institut Pasteur de Côte d’Ivoire expressed satisfaction with the entire programme; “The theme was relevant, the course was easy to access, the schedule considered the main topics. The instructors were good and available to answer questions. The number of participants allowed the instructors to take care of us during practical sessions. Although the duration of the course was not adequate for us, the beginners, to learn more about Linux, R and bioinformatics, it was a good experience overall and I will recommend it to people around me”, he said. Another participant, Issah Samori, a Graduate Intern at WACCBIP said “the course made me learn more about helminths genomics and the power of bioinformatics tools in our everyday lives.”.